Abstract
Introduction Allogeneic hematopoietic stem cell transplantation (allo-HSCT) is a curative treatment for patients with hematologic malignances. Immune system genes like cytokines play a role in inflammatory process that occur during graft versus host disease (GVHD). Therefore, the analysis of polymorphisms that affect the expression or activity of these genes could be used as biomarkers to predict the development of complications after allo-HSCT. The aim of this study was to identify new polymorphisms in genes related to the immune response, mainly cytokines, that could be related to the development of post-transplant complications.
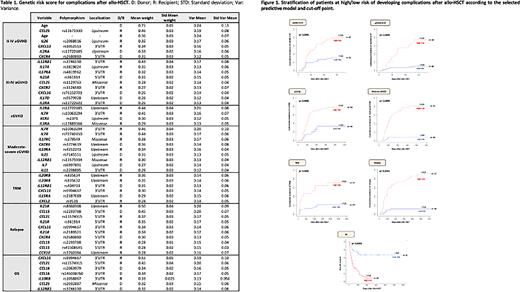
Methods We retrospectively selected 90 patients with hematological malignancies who received an allo-HSCT from an HLA-identical sibling donor from 2000 to 2015. The genotyping was performed using a custom NGS panel (73 interleukins and 59 chemokines genes) in a HiSeq platform (Illumina, USA). The bioinformatics analysis was carried out with the GeneSystems software (Sistemas Genómicos, Spain). We analyzed variants located in coding regions, splicing sites, UTR, and 5’ upstream and 3'downstream regions (+/-200pb). The polymorphisms selected corresponding to non-synonymous variants, with depth ≥30X in the canonical isoform with an allele frequency ≥0.4, with a minor allele frequency ≥1% and represented in at least 5% in our cohort. Bayesian logistic regression models (BLR) were used to select the most relevant variables for each post-transplant complication. The models with the highest AUC value, sensitivity and specificity value and the lowest number of genetic variants used were selected. Finally, the score obtained from the chosen predictive model for each complication was used to classify patients as low or high risk according to the selected cutoff point.
Results The cumulative incidence (CI) rates for aGVHD of grades II-IV and III-IV at 100 days after transplantation were 56.7% and 23.3%, respectively. The CI for cGVHD and moderate-severe cGVHD was 43.3% and 27.8%, respectively, at 3 years after allo-HSCT. The CI of TRM at 3 years was 24.4% and OS at 5 years was 30%.
Using filters defined previously, we detected 737 polymorphisms (644 in non-coding regions and 93 in coding regions). We developed BLR models for II-IV and III-IV aGVHD, cGVHD, moderate-severe cGVHD, TRM, relapse and OS (Table 1). Stratification of the cohort of patients according to the risk of develop each complication is showed in Figure 1.
Most polymorphisms found are located in regulatory regions, which until have been little studied, but these regions are involved in the expression genes, therefore could be affecting the function of these genes.
Conclusions We have identified new genetic polymorphisms correlate with the risk of developing complications after allo-HSCT from an HLA-identical sibling donor. Based on these data, we have developed a genetic score that encompasses polymorphisms of greater relevance. In this way, patients at greatest risk for developing post-transplant complication who could benefit from personalized management through immunosuppression and other drugs.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.